Research
We conduct research to bridge the gap between life sciences and computer science, employing advanced sequencing technologies to analyze (meta-)genomic and transcriptomic datasets related to infectious diseases, immunology, and medical sciences. With this interdisciplinary approach, we aim to improve our understanding of pathogen-host interactions and support genomic surveillance and containment.
Our research currently focuses on metagenomics, pangenomics, and wastewater surveillance in the context of infectious diseases and public health improvement.
Highlighted
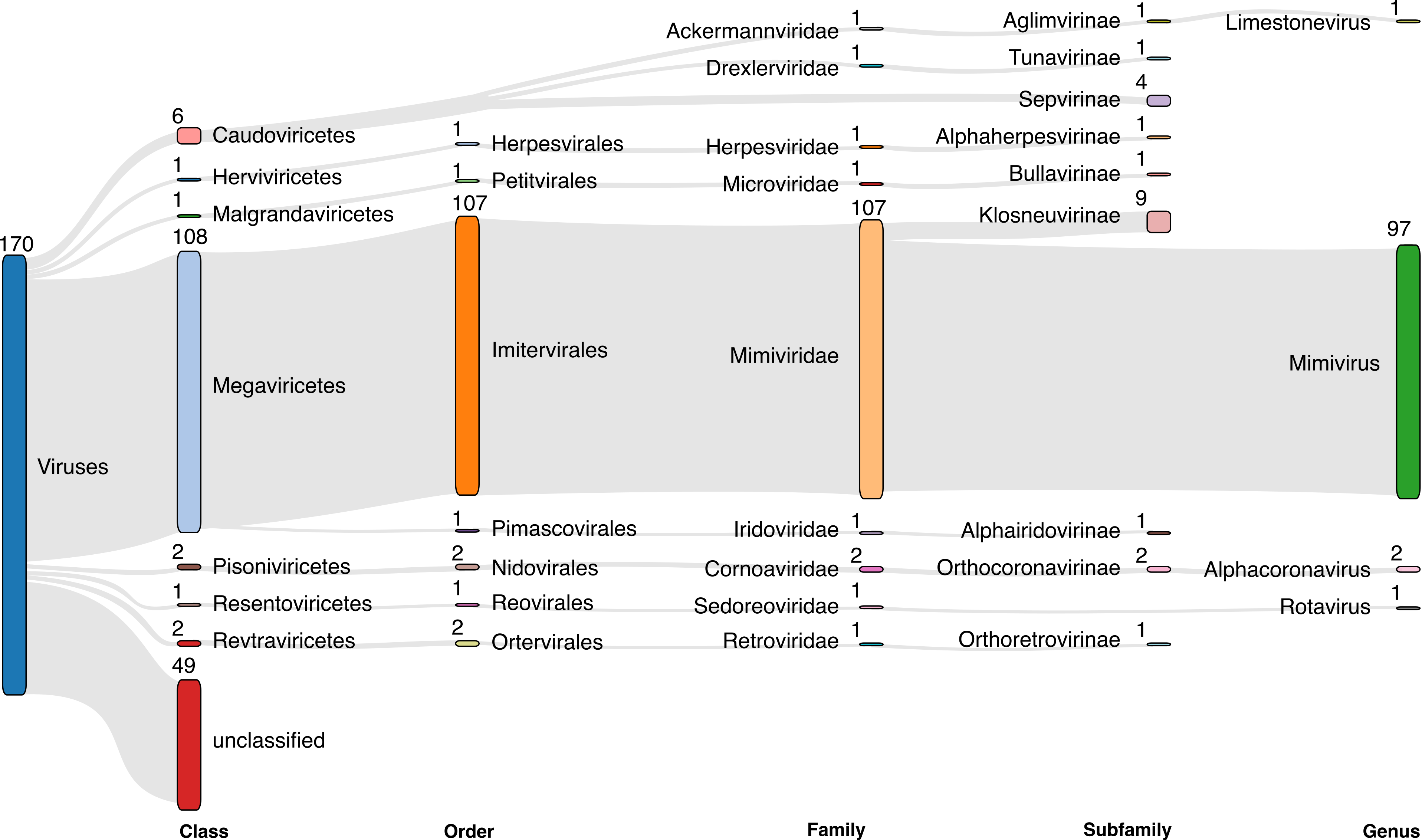
VIRify: An integrated detection, annotation and taxonomic classification pipeline using virus-specific protein profile hidden Markov models
PLOS Computational Biology
·
28 Aug 2023
·
doi:10.1371/journal.pcbi.1011422

Impact of reference design on estimating SARS-CoV-2 lineage abundances from wastewater sequencing data
Cold Spring Harbor Laboratory
·
02 Jun 2023
·
doi:10.1101/2023.06.02.543047
All
Note: this list currently includes all publications by lab members, including work they may have published while they were in previous labs.
2024
Reannotation of the zebra finch genome reveals undiscovered transcriptional complexity
Cold Spring Harbor Laboratory
·
09 Jan 2024
·
doi:10.1101/2024.01.09.574630
2023
Dicer‐dependent heterochromatic small RNAs in the model diatom species Phaeodactylum tricornutum
New Phytologist
·
03 Dec 2023
·
doi:10.1111/nph.19429
decOM: similarity-based microbial source tracking of ancient oral samples using k-mer-based methods
Microbiome
·
06 Nov 2023
·
doi:10.1186/s40168-023-01670-3
aKmerBroom: Ancient oral DNA decontamination using Bloom filters on k-mer sets
iScience
·
01 Nov 2023
·
doi:10.1016/j.isci.2023.108057
Refphase: Multi-sample phasing reveals haplotype-specific copy number heterogeneity
PLOS Computational Biology
·
23 Oct 2023
·
doi:10.1371/journal.pcbi.1011379
Lessons learned: overcoming common challenges in reconstructing the SARS-CoV-2 genome from short-read sequencing data via CoVpipe2
F1000Research
·
01 Sep 2023
·
doi:10.12688/f1000research.136683.1
VIRify: An integrated detection, annotation and taxonomic classification pipeline using virus-specific protein profile hidden Markov models
PLOS Computational Biology
·
28 Aug 2023
·
doi:10.1371/journal.pcbi.1011422
Targeted decontamination of sequencing data with CLEAN
Cold Spring Harbor Laboratory
·
06 Aug 2023
·
doi:10.1101/2023.08.05.552089
Impact of reference design on estimating SARS-CoV-2 lineage abundances from wastewater sequencing data
Cold Spring Harbor Laboratory
·
02 Jun 2023
·
doi:10.1101/2023.06.02.543047
Pangenome calculation beyond the species level using RIBAP: A comprehensive bacterial core genome annotation pipeline based on Roary and pairwise ILPs
Cold Spring Harbor Laboratory
·
07 May 2023
·
doi:10.1101/2023.05.05.539552
Building alternative splicing and evolution-aware sequence-structure maps for protein repeats
Cold Spring Harbor Laboratory
·
30 Apr 2023
·
doi:10.1101/2023.04.29.538821
De novogenome assembly resolving repetitive structures enables genomic analysis of 35 EuropeanMycoplasma bovisstrains
Cold Spring Harbor Laboratory
·
13 Apr 2023
·
doi:10.1101/2023.04.13.536562
Sequencing accuracy and systematic errors of nanopore direct RNA sequencing
Cold Spring Harbor Laboratory
·
30 Mar 2023
·
doi:10.1101/2023.03.29.534691
Nanopore-based enrichment of antimicrobial resistance genes – a case-based study
Gigabyte
·
25 Jan 2023
·
doi:10.46471/gigabyte.75
Managing and monitoring a pandemic: showcasing a practical approach for the genomic surveillance of SARS-CoV-2
Database
·
01 Jan 2023
·
doi:10.1093/database/baad071
2022
Refphase: Multi-sample reference phasing reveals haplotype-specific copy number heterogeneity
Cold Spring Harbor Laboratory
·
17 Oct 2022
·
doi:10.1101/2022.10.13.511885
VIRify: an integrated detection, annotation and taxonomic classification pipeline using virus-specific protein profile hidden Markov models
Cold Spring Harbor Laboratory
·
22 Aug 2022
·
doi:10.1101/2022.08.22.504484
CovRadar: continuously tracking and filtering SARS-CoV-2 mutations for genomic surveillance
Bioinformatics
·
07 Jul 2022
·
doi:10.1093/bioinformatics/btac411
DiNAMO: Exact method for degenerate IUPAC motifs discovery, characterization of sequence-specific errors
[no publisher info]
·
07 Jun 2022
·
hal:hal-01574630
Assessing Conservation of Alternative Splicing with Evolutionary Splicing Graphs
Genome Research
·
07 Jun 2022
·
hal:hal-03274456
Statistical methods for analysing high throughput sequencing data
[no publisher info]
·
07 Jun 2022
·
hal:tel-03636135
Identification of programmed translational -1 frameshifting sites in the genome of Saccharomyces cerevisiae.
Genome research
·
30 May 2022
·
pmid:16882864
Role of the chromatin landscape and sequence in determining cell type-specific genomic glucocorticoid receptor binding and gene regulation.
Nucleic acids research
·
25 May 2022
·
pmid:27903902
Near Chromosome-Level Genome Assembly and Annotation of Rhodotorula babjevae Strains Reveals High Intraspecific Divergence
Journal of Fungi
·
22 Mar 2022
·
doi:10.3390/jof8040323
ASES: visualizing evolutionary conservation of alternative splicing in proteins
Bioinformatics
·
21 Feb 2022
·
doi:10.1093/bioinformatics/btac105
Investigation of a Limited but Explosive COVID-19 Outbreak in a German Secondary School
Viruses
·
04 Jan 2022
·
doi:10.3390/v14010087
What the Phage: a scalable workflow for the identification and analysis of phage sequences
GigaScience
·
01 Jan 2022
·
doi:10.1093/gigascience/giac110
2021
Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions
Nature Microbiology
·
24 Dec 2021
·
doi:10.1038/s41564-021-01011-w
Context-aware genomic surveillance reveals hidden transmission of a carbapenemase-producing Klebsiella pneumoniae
Microbial Genomics
·
24 Dec 2021
·
doi:10.1099/mgen.0.000741
Spatial and temporal intratumour heterogeneity has potential consequences for single biopsy-based neuroblastoma treatment decisions
Nature Communications
·
23 Nov 2021
·
doi:10.1038/s41467-021-26870-z
Comparative transcriptomics identifies the transcription factors BRANCHED1 and TCP4, as well as the microRNA miR166 as candidate genes involved in the evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae)
Cold Spring Harbor Laboratory
·
01 Nov 2021
·
doi:10.1101/2021.10.29.466389
Mutations in the gdpP gene are a clinically relevant mechanism for β-lactam resistance in meticillin-resistant Staphylococcus aureus lacking mec determinants
Microbial Genomics
·
09 Sep 2021
·
doi:10.1099/mgen.0.000623
Rise and Fall of SARS-CoV-2 Lineage A.27 in Germany
Viruses
·
29 Jul 2021
·
doi:10.3390/v13081491
The economical lifestyle of CPR bacteria in groundwater allows little preference for environmental drivers
Cold Spring Harbor Laboratory
·
29 Jul 2021
·
doi:10.1101/2021.07.28.454184
Evidence for the existence of a new genus Chlamydiifrater gen. nov. inside the family Chlamydiaceae with two new species isolated from flamingo (Phoenicopterus roseus): Chlamydiifrater phoenicopteri sp. nov. and Chlamydiifrater volucris sp. nov.
Systematic and Applied Microbiology
·
01 Jul 2021
·
doi:10.1016/j.syapm.2021.126200
Assessing conservation of alternative splicing with evolutionary splicing graphs
Genome Research
·
15 Jun 2021
·
doi:10.1101/gr.274696.120
A global metagenomic map of urban microbiomes and antimicrobial resistance
Cell
·
01 Jun 2021
·
doi:10.1016/j.cell.2021.05.002
poreCov - an easy to use, fast, and robust workflow for SARS-CoV-2 genome reconstruction via nanopore sequencing
Cold Spring Harbor Laboratory
·
07 May 2021
·
doi:10.1101/2021.05.07.443089
Metagenomics workflow for hybrid assembly, differential coverage binning, metatranscriptomics and pathway analysis (MUFFIN)
PLOS Computational Biology
·
09 Feb 2021
·
doi:10.1371/journal.pcbi.1008716
RUNX3 Transcript Variants Have Distinct Roles in Ovarian Carcinoma and Differently Influence Platinum Sensitivity and Angiogenesis
Cancers
·
26 Jan 2021
·
doi:10.3390/cancers13030476
SLC35F2, a Transporter Sporadically Mutated in the Untranslated Region, Promotes Growth, Migration, and Invasion of Bladder Cancer Cells
Cells
·
06 Jan 2021
·
doi:10.3390/cells10010080
2020
RNAflow: An Effective and Simple RNA-Seq Differential Gene Expression Pipeline Using Nextflow
Genes
·
10 Dec 2020
·
doi:10.3390/genes11121487
A marine Chlamydomonas sp. emerging as an algal model
Journal of Phycology
·
22 Nov 2020
·
doi:10.1111/jpy.13083
Assessing Conservation of Alternative Splicing with Evolutionary Splicing Graphs
Cold Spring Harbor Laboratory
·
15 Nov 2020
·
doi:10.1101/2020.11.14.382820
A decade of de novo transcriptome assembly: Are we there yet?
Molecular Ecology Resources
·
29 Oct 2020
·
doi:10.1111/1755-0998.13268
Comparative Genome Analysis of 33 Chlamydia Strains Reveals Characteristic Features of Chlamydia Psittaci and Closely Related Species
Pathogens
·
28 Oct 2020
·
doi:10.3390/pathogens9110899
Inclusion of Oxford Nanopore long reads improves all microbial and viral metagenome‐assembled genomes from a complex aquifer system
Environmental Microbiology
·
20 Aug 2020
·
doi:10.1111/1462-2920.15186
What the Phage: A scalable workflow for the identification and analysis of phage sequences
Cold Spring Harbor Laboratory
·
25 Jul 2020
·
doi:10.1101/2020.07.24.219899
Computational Strategies to Combat COVID-19: Useful Tools to Accelerate SARS-CoV-2 and Coronavirus Research
MDPI AG
·
23 May 2020
·
doi:10.20944/preprints202005.0376.v1
PoSeiDon: a Nextflow pipeline for the detection of evolutionary recombination events and positive selection
Cold Spring Harbor Laboratory
·
20 May 2020
·
doi:10.1101/2020.05.18.102731
EpiDope: A Deep neural network for linear B-cell epitope prediction
Cold Spring Harbor Laboratory
·
13 May 2020
·
doi:10.1101/2020.05.12.090019
Targeted domain assembly for fast functional profiling of metagenomic datasets with S3A
Bioinformatics
·
24 Apr 2020
·
doi:10.1093/bioinformatics/btaa272
Transcripts’ Evolutionary History and Structural Dynamics Give Mechanistic Insights into the Functional Diversity of the JNK Family
Journal of Molecular Biology
·
01 Mar 2020
·
doi:10.1016/j.jmb.2020.01.032
Metagenomics workflow for hybrid assembly, differential coverage binning, transcriptomics and pathway analysis (MUFFIN)
Cold Spring Harbor Laboratory
·
08 Feb 2020
·
doi:10.1101/2020.02.08.939843
2019
Inclusion of Oxford Nanopore long reads improves all microbial and phage metagenome-assembled genomes from a complex aquifer system
Cold Spring Harbor Laboratory
·
19 Dec 2019
·
doi:10.1101/2019.12.18.880807
Addressing dereplication crisis: Taxonomy-free reduction of massive genome collections using embeddings of protein content
Cold Spring Harbor Laboratory
·
25 Nov 2019
·
doi:10.1101/855262
Chlamydia buteonis, a new Chlamydia species isolated from a red-shouldered hawk
Systematic and Applied Microbiology
·
01 Sep 2019
·
doi:10.1016/j.syapm.2019.06.002
Virus- and Interferon Alpha-Induced Transcriptomes of Cells from the Microbat Myotis daubentonii
iScience
·
01 Sep 2019
·
doi:10.1016/j.isci.2019.08.016
A comprehensive annotation and differential expression analysis of short and long non-coding RNAs in 16 bat genomes
Cold Spring Harbor Laboratory
·
19 Aug 2019
·
doi:10.1101/738526
Global Genetic Cartography of Urban Metagenomes and Anti-Microbial Resistance
Cold Spring Harbor Laboratory
·
05 Aug 2019
·
doi:10.1101/724526
Evaluation of Sequencing Library Preparation Protocols for Viral Metagenomic Analysis from Pristine Aquifer Groundwaters
Viruses
·
28 May 2019
·
doi:10.3390/v11060484
Structure and Hierarchy of Influenza Virus Models Revealed by Reaction Network Analysis
Viruses
·
16 May 2019
·
doi:10.3390/v11050449
De novo transcriptome assembly: A comprehensive cross-species comparison of short-read RNA-Seq assemblers
GigaScience
·
01 May 2019
·
doi:10.1093/gigascience/giz039
The Anti-amyloid Compound DO1 Decreases Plaque Pathology and Neuroinflammation-Related Expression Changes in 5xFAD Transgenic Mice
Cell Chemical Biology
·
01 Jan 2019
·
doi:10.1016/j.chembiol.2018.10.013
SilentMutations (SIM): A tool for analyzing long-range RNA–RNA interactions in viral genomes and structured RNAs
Virus Research
·
01 Jan 2019
·
doi:10.1016/j.virusres.2018.11.005
2018
Cohesin-mediated NF-κB signaling limits hematopoietic stem cell self-renewal in aging and inflammation
Journal of Experimental Medicine
·
07 Dec 2018
·
doi:10.1084/jem.20181505
Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis
Cold Spring Harbor Laboratory
·
30 Nov 2018
·
doi:10.1101/483693
PCAGO: An interactive web service to analyze RNA-Seq data with principal component analysis
Cold Spring Harbor Laboratory
·
03 Oct 2018
·
doi:10.1101/433078
DiNAMO: highly sensitive DNA motif discovery in high-throughput sequencing data
BMC Bioinformatics
·
11 Jun 2018
·
doi:10.1186/s12859-018-2215-1
Meet-U: Educating through research immersion
PLOS Computational Biology
·
15 Mar 2018
·
doi:10.1371/journal.pcbi.1005992
2017
PureCLIP: capturing target-specific protein–RNA interaction footprints from single-nucleotide CLIP-seq data
Genome Biology
·
01 Dec 2017
·
doi:10.1186/s13059-017-1364-2
Jointly aligning a group of DNA reads improves accuracy of identifying large deletions
Nucleic Acids Research
·
22 Nov 2017
·
doi:10.1093/nar/gkx1175
Comprehensive insights in the Mycobacterium avium subsp. paratuberculosis genome using new WGS data of sheep strain JIII-386 from Germany.
Genome biology and evolution
·
24 Oct 2017
·
pmc:PMC4607514
Complete Genome Sequence of JII-1961, a Bovine Mycobacterium avium subsp.
paratuberculosis
Field Isolate from Germany
Genome Announcements
·
24 Aug 2017
·
doi:10.1128/genomea.00870-17
Evolution and Antiviral Specificities of Interferon-Induced Mx Proteins of Bats against Ebola, Influenza, and Other RNA Viruses
Journal of Virology
·
01 Aug 2017
·
doi:10.1128/jvi.00361-17
PureCLIP: capturing target-specific protein-RNA interaction footprints from single-nucleotide CLIP-seq data
Cold Spring Harbor Laboratory
·
07 Jun 2017
·
doi:10.1101/146704
Transcripts' evolutionary history and structural dynamics give mechanistic insights into the functional diversity of the JNK family.
Cold Spring Harbor Laboratory
·
23 Mar 2017
·
doi:10.1101/119891
Evaluation of associations between genotypes of Mycobacterium avium subsp. paratuberculsis and presence of intestinal lesions characteristic of paratuberculosis
Veterinary Microbiology
·
01 Mar 2017
·
doi:10.1016/j.vetmic.2017.01.026
Massive Effect on LncRNAs in Human Monocytes During Fungal and Bacterial Infections and in Response to Vitamins A and D
Scientific Reports
·
17 Jan 2017
·
doi:10.1038/srep40598
Differential Effects of Vitamins A and D on the Transcriptional Landscape of Human Monocytes during Infection
Scientific Reports
·
17 Jan 2017
·
doi:10.1038/srep40599
Software Dedicated to Virus Sequence Analysis “Bioinformatics Goes Viral”
In Loeffler’s Footsteps – Viral Genomics in the Era of High-Throughput Sequencing
·
01 Jan 2017
·
doi:10.1016/bs.aivir.2017.08.004
2016
Improved Prediction of Non-methylated Islands in Vertebrates Highlights Different Characteristic Sequence Patterns
PLOS Computational Biology
·
16 Dec 2016
·
doi:10.1371/journal.pcbi.1005249
Differential transcriptional responses to Ebola and Marburg virus infection in bat and human cells
Scientific Reports
·
07 Oct 2016
·
doi:10.1038/srep34589
Predicting enhancers using a small subset of high confidence examples and co-training
PeerJ
·
01 Sep 2016
·
doi:10.7287/peerj.preprints.2407
Predicting enhancers using a small subset of high confidence examples and co-training
PeerJ
·
01 Sep 2016
·
doi:10.7287/peerj.preprints.2407v1
Whole-Genome Sequence of
Chlamydia gallinacea
Type Strain 08-1274/3
Genome Announcements
·
25 Aug 2016
·
doi:10.1128/genomea.00708-16
The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International Consortium inaugural meeting report
Microbiome
·
03 Jun 2016
·
doi:10.1186/s40168-016-0168-z
2015
SPON2, a newly identified target gene of MACC1, drives colorectal cancer metastasis in mice and is prognostic for colorectal cancer patient survival
Oncogene
·
21 Dec 2015
·
doi:10.1038/onc.2015.451
Towards a comprehensive picture of alloacceptor tRNA remolding in metazoan mitochondrial genomes
Nucleic Acids Research
·
30 Jul 2015
·
doi:10.1093/nar/gkv746
X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes
Molecular Psychiatry
·
03 Feb 2015
·
doi:10.1038/mp.2014.193
2014
Ulysses: accurate detection of low-frequency structural variations in large insert-size sequencing libraries
Bioinformatics
·
07 Nov 2014
·
doi:10.1093/bioinformatics/btu730
Fiona: a parallel and automatic strategy for read error correction
Bioinformatics
·
22 Aug 2014
·
doi:10.1093/bioinformatics/btu440
The diversity of small non-coding RNAs in the diatom Phaeodactylum tricornutum
BMC Genomics
·
20 Aug 2014
·
doi:10.1186/1471-2164-15-698
Concerted down-regulation of immune-system related genes predicts metastasis in colorectal carcinoma
BMC Cancer
·
05 Feb 2014
·
doi:10.1186/1471-2407-14-64
Parseq: reconstruction of microbial transcription landscape from RNA-Seq read counts using state-space models
Bioinformatics
·
27 Jan 2014
·
doi:10.1093/bioinformatics/btu042
2013
Assessment of transcript reconstruction methods for RNA-seq
Nature Methods
·
03 Nov 2013
·
doi:10.1038/nmeth.2714
PROmiRNA: a new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs
Genome Biology
·
01 Jan 2013
·
doi:10.1186/gb-2013-14-8-r84
2011
Comprehensive identification and quantification of microbial transcriptomes by genome-wide unbiased methods
Current Opinion in Biotechnology
·
01 Feb 2011
·
doi:10.1016/j.copbio.2010.10.003
2010
The soluble intracellular domain of megalin does not affect renal proximal tubular function in vivo
Kidney International
·
01 Sep 2010
·
doi:10.1038/ki.2010.169
The genome of a songbird
Nature
·
01 Apr 2010
·
doi:10.1038/nature08819
Prediction of alternative isoforms from exon expression levels in RNA-Seq experiments
Nucleic Acids Research
·
11 Feb 2010
·
doi:10.1093/nar/gkq041
2009
Mutation screening in 86 known X-linked mental retardation genes by droplet-based multiplex PCR and massive parallel sequencing
The HUGO Journal
·
01 Dec 2009
·
doi:10.1007/s11568-010-9137-y
Erratum to: Mutation screening in 86 known X-linked mental retardation genes by droplet-based multiplex PCR and massive parallel sequencing
The HUGO Journal
·
01 Dec 2009
·
doi:10.1007/s11568-010-9142-1
DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction
Nature Genetics
·
04 Oct 2009
·
doi:10.1038/ng.463
MedlineRanker: flexible ranking of biomedical literature
Nucleic Acids Research
·
07 May 2009
·
doi:10.1093/nar/gkp353
Detection of Alpha-Rod Protein Repeats Using a Neural Network and Application to Huntingtin
PLoS Computational Biology
·
13 Mar 2009
·
doi:10.1371/journal.pcbi.1000304
Recent developments in StemBase: a tool to study gene expression in human and murine stem cells
BMC Research Notes
·
01 Jan 2009
·
doi:10.1186/1756-0500-2-39
2008
A Global View of Gene Activity and Alternative Splicing by Deep Sequencing of the Human Transcriptome
Science
·
15 Aug 2008
·
doi:10.1126/science.1160342
2007
BiasViz: visualization of amino acid biased regions in protein alignments
Bioinformatics
·
06 Oct 2007
·
doi:10.1093/bioinformatics/btm489
2005
Identification of programmed translational -1 frameshifting sites in the genome of Saccharomyces cerevisiae
Genome Research
·
01 Oct 2005
·
doi:10.1101/gr.4258005
seq++: analyzing biological sequences with a range of Markov-related models
Bioinformatics
·
17 Mar 2005
·
doi:10.1093/bioinformatics/bti389
Study of stem cell function using microarray experiments
FEBS Letters
·
23 Feb 2005
·
doi:10.1016/j.febslet.2005.02.020
2003
SPA: simple web tool to assess statistical significance of DNA patterns
Nucleic Acids Research
·
01 Jul 2003
·
doi:10.1093/nar/gkg613
SIC: a tool to detect short inverted segments in a biological sequence
Nucleic Acids Research
·
01 Jul 2003
·
doi:10.1093/nar/gkg596
2002
Occurrence Probability of Structured Motifs in Random Sequences
Journal of Computational Biology
·
01 Dec 2002
·
doi:10.1089/10665270260518254