Software
It is important to us that our bioinformatics research also leads to robust, easy-to-use and useful tools for the scientific community. Our tools are available open source and we use workflow management systems like Nextflow and containers like Docker to develop reproducible research software. Have fun trying them out and we’re always happy to receive feedback and pull requests on GitHub.
Featured
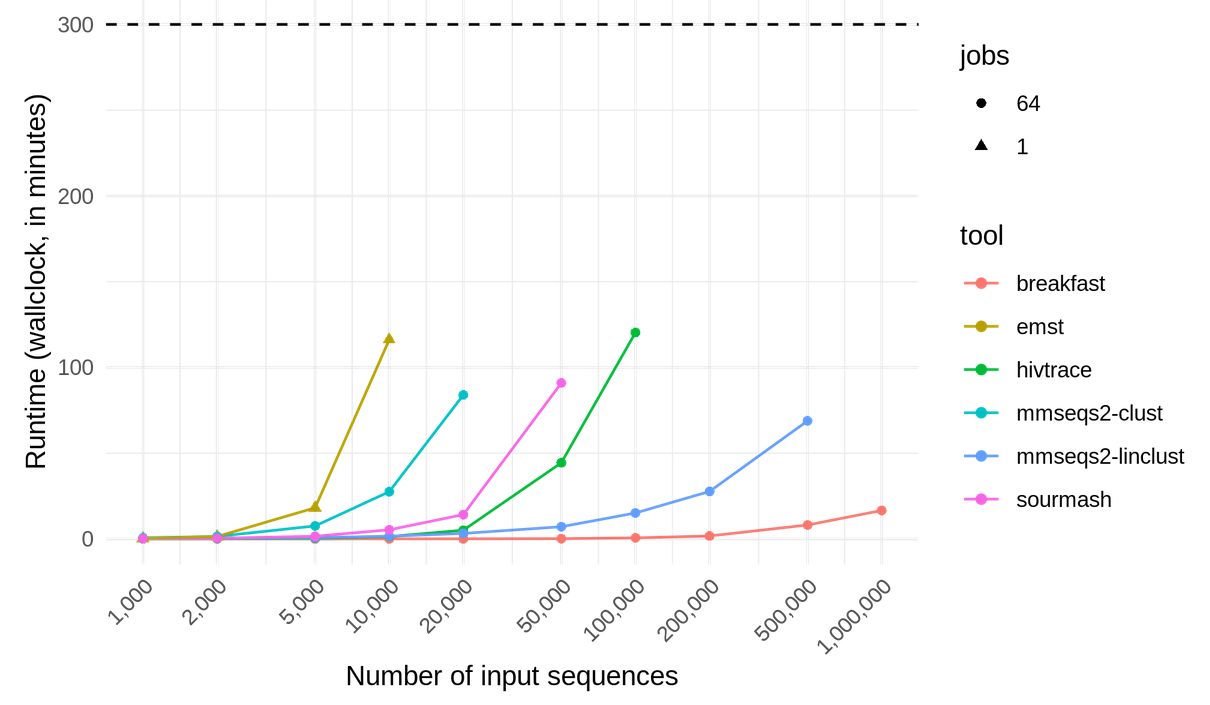
Fast detection of putative transmission chains in large (>1 million) sets of SARS-CoV-2 genomic sequences.
A comprehensive bacterial core genome annotation pipeline based on Roary and pairwise Integer Linear Programming. Calculate core genes beyond species levels.
More
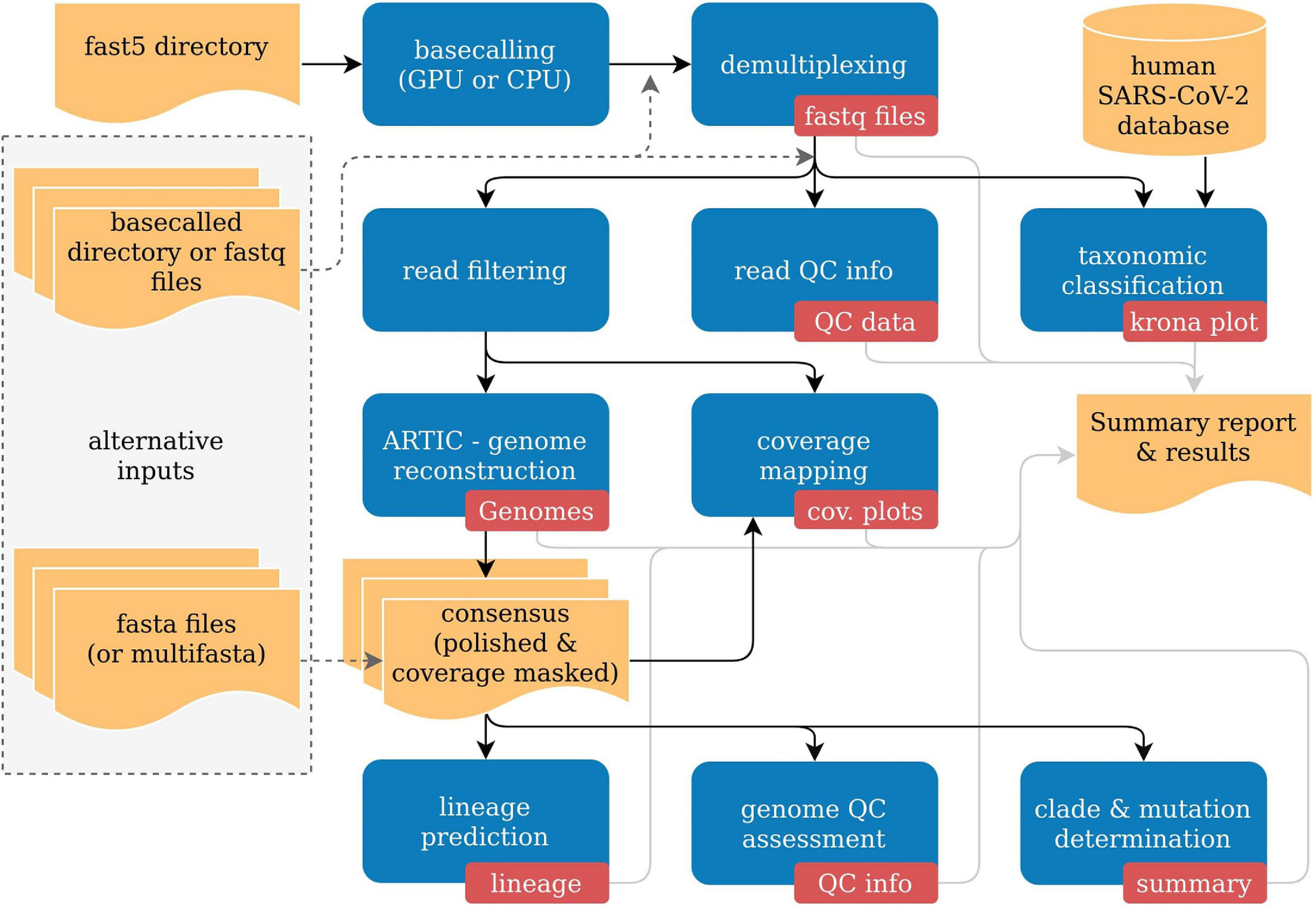
An easy to use, fast, and robust Nextflow pipeline for SARS-CoV-2 genome reconstruction from Nanopore sequencing data. Developed together w/ the CaSe Group.

Detection of phages and eukaryotic viruses from metagenomic and metatranscriptomic assemblies. Developed together and mainly maintained by the Microbiome Informatics team at EBI.

A portable, scalable, and parallelizable Nextflow RNA-Seq pipeline to detect differentially expressed genes, aiming for a high level of reproducibility.
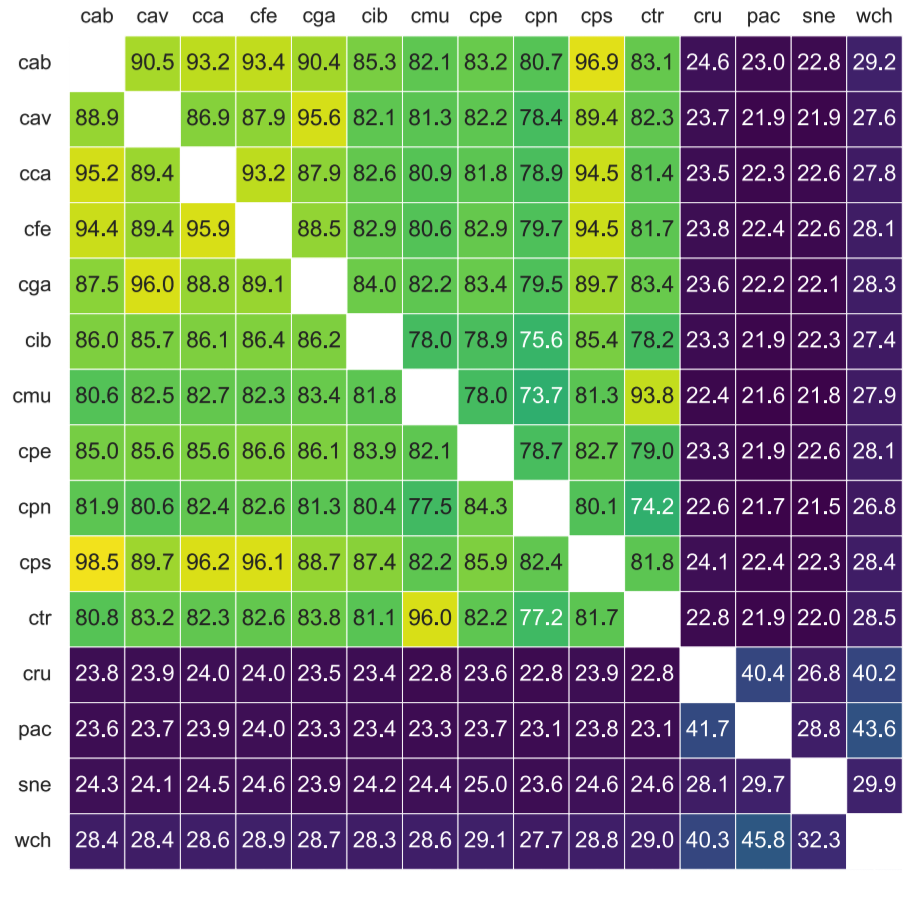
Nextflow pipeline for the Calculation of the Percentage Of Conserved Proteins following Qin et al. 2014. but using DIAMOND instead of BLASTP.

A pipeline for Positive Selection Detection and recombination analysis of protein-coding genes.
Interactive web service that allows analysis of RNA-Seq read count data with PCA and clustering. Go to web service. Run desktop app.